
|
Program Overview > Program Members > Atsushi, Koito
|

Faculty of Life Sciences, Professor
Virology, M.D., Ph.D. Member of International Research Committee, Research on the molecular basis of HIV infection

 |
 HIV exhibits a highly restricted host cell tropism. Transgenic rodents whose CD4+ T cells could support HIV infection, have been expect to prove useful as a small-animal model, however, exhibited little or no sign of productive infection (1). We found that the Gag fails to be targeted to the plasma membrane that results in failure of HIV to spread in rodent cells (2, 3). Recently, powerful intracellular defense mechanism(s) against the retroviral infectivity, mobility of endogenous retroelements and other viruses have been identified from the studies on the host restriction factor(s) against HIV replication. In addition to the conventional innate and acquired immune responses, vertebrate lineage, especially mammalian species are found to have evolved a series of dominant, constitutively expressed genes that inhibit infection by retroviruses. |
|
The early block owing to Fv1 and TRIM5α that target incoming retroviral capsids has been shown to be one of the major restriction factors against retrovirus infection. In addition, the APOBEC3 family of the cytidine deaminases at the late phase that hypermutate and disrupt the retroviral genomes, has emerged as an important class of innate antiretroviral resistance factors. We are currently attempting to clarify the molecular basis of retrovirus restriction activities such as APOBECs possessed by a variety of mammalian species. There is also now evidence that some APOBEC3 proteins can target a variety of retroviral substrates, such as various oncovirus, whereas HTLV-1 has been shown to be relatively resistant to these citidine deaminases (4). Although the role of the APOBEC3-dependent retroelement restriction system as an intrinsic resistance mechanism is becoming clear, less well understood is mammalian APOBEC1, the catalytic component of a complex that deaminates apolipoprotein B (apoB) mRNA in gastrointestinal tissue. We have identified the several APOBEC1s from small animal species can suppress HIV replication mainly through deaminase-dependent mechanism, whereas human APOBEC1 had negligible effects. Interestingly, these APOBEC1s, which act on both first-strand DNA and genomic RNA in the virions, showed a clear WCW (W is A or U) trinucleotide preference that was reported to be highly conserved as the apoB mRNA editing site sequence by APOBEC1s among divergent of mammalian species (5). These results indicate that APOBEC1 may function as the defense mechanism regulating various retroelements and invading nucleic acids in a wide range of mammalian species. Understanding these species-specific or non-specific repressive activities to HIV may suggest approaches to development of small-animal model of HIV infection. In addition, the elucidation of these repressive activities may lead to development of a novel therapeutic intervention in retroviral infection. |
|
|
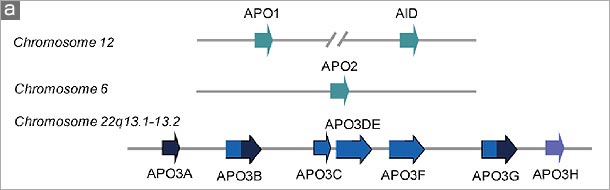
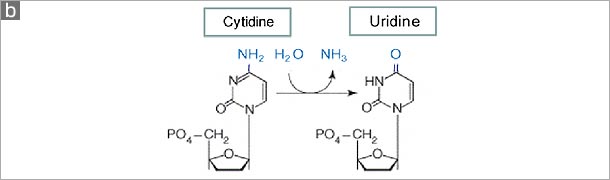
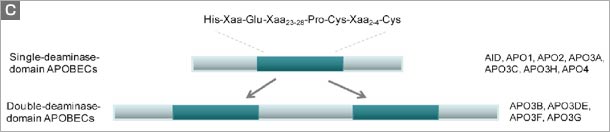
Figure. The human APOBEC family. (a) A schematic of the human genome contains members of the AID/APOBEC family. There are several forms of mammalian AID/APOBEC proteins with distinct functions in vivo: activation-induced deaminase (AID), APOBEC1 (APO1), APOBEC2 (APO2), APOBEC3 (APO3) and APOBEC4 (APO4). The primate-specific cluster of at least seven APO3 related genes: APO3A-3H, resides on the same chromosome within ~150 kb. In contrast, mice and rats retain only a single APO3 gene, located on chromosome 15e2 and 7q34 respectively, indicating a relatively recent, and possibly unprecedented gene expansion has occurred in mammalian species. APO1 and AID are located approximately 900 kb apart on human chromosome 12. (b) The cytidine deamination reaction catalysed by APO3 is similar to that of APO1 and AID. In this process, cytidine is converted to uridine by the addition of water molecule (H2O) and removal of an ammonia (NH3). (c) The AID/APOBEC family proteins, of which APO3 is a member, share one or two copies of the active site core motif His-Xaa-Glu-Xaa23-28-Pro-Cys-Xaa2-4-Cys (where X denotes any amino acid) in the cytidine deaminase domain. The histidine (His) and the two cysteine (Cys) residues within the core coordinate Zn2+ ion, and a glutamamic acid (Glu) serves an essential role in catalysis as a proton shuttle in the deamination reaction . |

|
♦Reference |
|
| 1. | Sawada, Gowrishankar, Kitamura, Suzuki, Suzuki, Tahara & Koito.: J. Exp. Med. 187: 1439, 1998. |
| 2. | Koito, Shigekane & Matsushita.: Virology 305:181, 2003. |
| 3. | Koito, Kameyama, Cheng-Mayer & Matsushita.: J. Virol. 77: 5109, 2003. |
| 4. | Ohsugi & Koito.: J. Virol. Method 139: 93, 2007. |
| 5. | Ikeda, Ohsugi, Kimura, Matsushita, Maeda, Harada & Koito.: Nucleic Acids Res. 36: 6859, 2008. |
 |
 |