Identification and characterization of a novel enhancer in the HTLV-1 proviral genome
Points
- Human T-cell leukemia virus type I (HTLV-1) is a retrovirus that is integrated to host genome and cause chronic persistent infection. It is extremely difficult to eliminate the virus from infected individuals because of their integration.
- In this study, we identified a novel transcription regulatory region (enhancer region) in the provirus that is important for the establishment of chronic persistent infection.
- In order to investigate the phenomena occurring in the bodies of infected individuals, this study was conducted using actual infected specimens and single-cell analysis techniques, which led to this new discovery.
- These findings are important for understanding the persistence and pathogenesis of HTLV-1 infection and will lead to the creation of new therapeutic targets to prevent chronic persistent HTLV-1 infection.
Research Summary
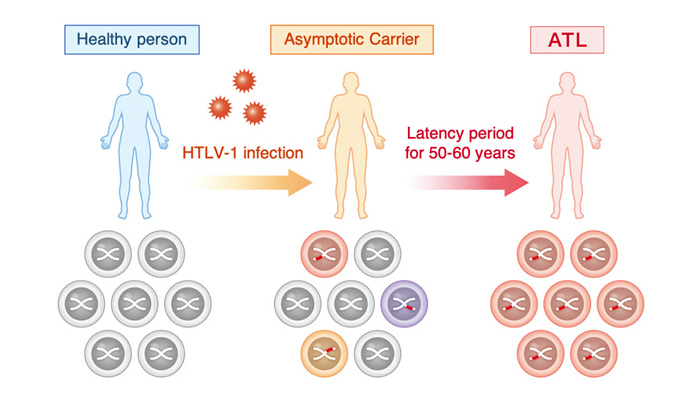
HTLV-1 is a retrovirus that is pathogenic to humans and primarily targets CD4+ T cells, which play a central role in the human immune system, for infection. After entering the host cell, the virus inserts viral genome into the host genome and is maintained as a provirus. Once infection is established, the infection becomes chronic and persistent, but the majority of infected individuals remain asymptomatic for the rest of their lives without developing any associated diseases. On the other hand, 2-5% of infected individuals develop adult T-cell leukemia (ATL) after a long latency period of approximately 60 years (Figure 1). ATL is known to be a very poor prognosis leukemia.
The host genomic DNA of ATL cells contains proviral DNA, and the presence of the provirus is thought to induce cellular oncogenesis, since CD4+ T cell cancers are extremely rare unless infected with HTLV-1. Therefore, although many studies have been conducted on the integrated provirus, the existence of the viral enhancer identified in this study was unknown for more than 40 years after the discovery of the virus.
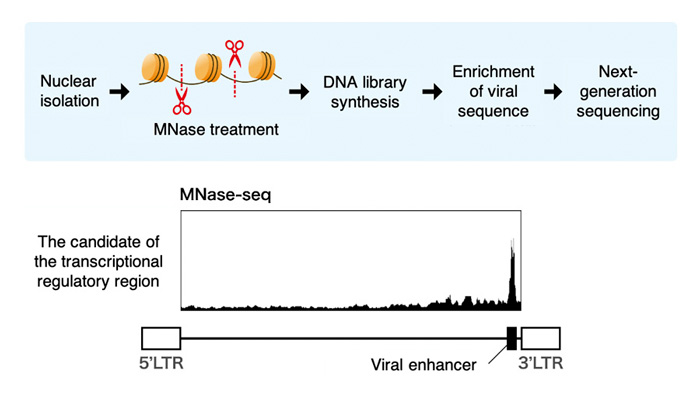
In collaboration with a medical institution in Kyushu, this group comprehensively investigated the transcriptional regulatory regions within the HTLV-1 provirus by performing MNase-seq using a highly sensitive viral sequencing method on samples from infected patients. As a result, a previously unidentified transcriptional regulatory region (enhancer region) in the viral genome was identified (Figure 2). In addition, the mechanism of the enhancer activity was investigated, and the involvement of two host cell factors, SRF and ELK-1, was clarified (Figure 3).
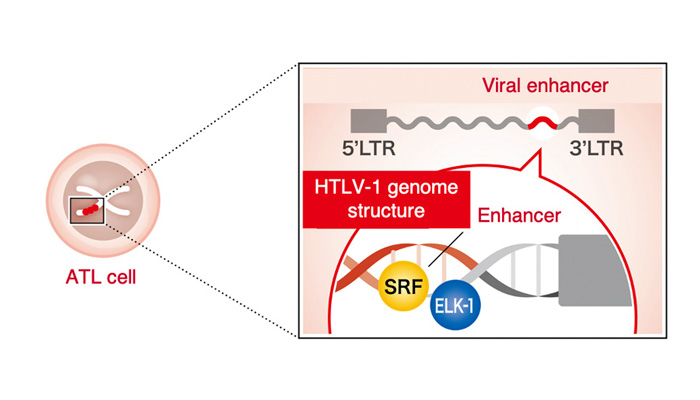
It is also known that host cell gene expression is activated around the integration sites of HTLV-1 provirus in the ATL cell genome, which has been reported to contribute to the development of leukemia. In this study, the HTLV-1 viral enhancer not only contributes to the maintenance of viral gene expression, but also affects on host gene expression around the integration sites.(Figure 4).
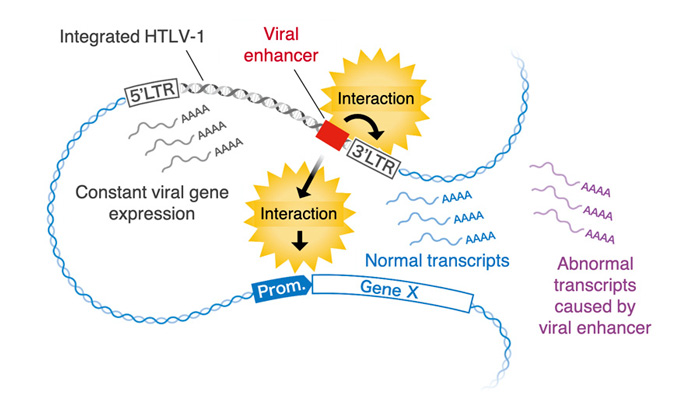
This study revealed the existence of a novel enhancer region in the HTLV-1 provirus and that function, which could not be found by conventional methods, by analyzing patient samples using advanced research methods such as next-generation sequencing and single cell analysis. The results of this study clarify one aspect of the mechanism of persistence and pathogenicity of HTLV-1 infection, and that will lead to a step overcoming the problem of HTLV-1 infection.
Publication information
- Title:
- Identification and characterization of a novel enhancer in the HTLV-1 proviral genome
- Authors:
- Misaki Matsuo, Takaharu Ueno#, Kazuaki Monde#, Kenji Sugata#, Benjy Jek Yang Tan, Akhinur Rahman, Paola Miyazato, Kyosuke Uchiyama, Saiful Islam, Hiroo Katsuya, Shinsuke Nakajima, Masahito Tokunaga, Kisato Nosaka, Hiroyuki Hata, Atae Utsunomiya, Jun-ichi Fujisawa, Yorifumi Satou*
- Journal:
- Nature communications
- DOI:
- 10.1038/s41467-022-30029-9
- URL:
- https://www.nature.com/articles/s41467-022-30029-9
