Single cell immune profiling of peripheral blood mononuclear cells (PBMCs) in Human T-cell Leukemia Virus type 1 (HTLV-1) infected individuals
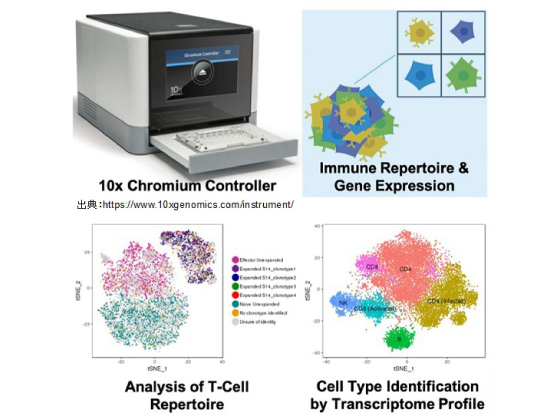
HTLV-1, although a distant relative of the Human Immunodeficiency Virus (HIV), causes disease in only 5-10% of infected people. Unlike HIV infection in which viremia needs to be contained with antiretroviral drugs, viremia in HTLV-1 infection is relatively well-controlled by the host immune response and only in a small population the infection progresses to chronic diseases such as adult T-cell leukemia (ATL) and HTLV-1 associated myelopathy/tropical spastic paraparesis (HAM/TSP). It is known that HTLV-1 dysregulates the host immune system; yet, despite extensive studies, how HTLV-1 induces disease remains unknown. Single cell technologies are now becoming increasingly important tools in biology and medicine. Single-cell measurements provide a finer look into complex biology and unmask cellular heterogeneity present in tissues to complement the information gleaned from bulk populations of cells. With the increasing sophistication of single-cell technology, a huge number of cells can now be analyzed to provide more information such as cellular subtype and clonal evolution. In our lab, we are currently using the Chromium 10x Genomics platform to perform single cell RNA-Seq (scRNA-Seq) on PBMCs from HTLV-1-infected individuals. Through this approach, we are hoping to obtain a finer insight into the expression profile of individual cells in order to identify factors which trigger progression of the disease from latent, asymptomatic state. Besides that, we are also studying the difference of expression profile between clonally expanded and non-expanded infected cells to discern their differences and understand what are the triggering factors for clonal expansion. We are thrilled with what can be performed using this advanced technology and we are looking forward to more exciting findings in the near future. Finally, this research project is part of the Japanese Initiative for Progress of Research on Infectious Disease for global Epidemic (J-PRIDE) of Japan Agency for Medical Research and Development (AMED).
